 |
United States Department of Agriculture - Agricultural Research Service |  |
| Northwest Irrigation & Soils Research Lab, Kimberly, ID | ||
Irrigated Agriculture Decreases Soil Microbial Genetic Diversity |
 |
United States Department of Agriculture - Agricultural Research Service |  |
| Northwest Irrigation & Soils Research Lab, Kimberly, ID | ||
Irrigated Agriculture Decreases Soil Microbial Genetic Diversity |
1USDA Agricultural Research Service, Northwest Irrigation and Soils Research Laboratory, Kimberly, ID
2Department of Biological Sciences & Department of Environmental Studies Florida International University, Miami, FL
Abstract
Increasing the amount of C in soils may be one method to reduce the concentration of CO2 in the atmosphere. We measured organic C stored in Southern Idaho soils having long term cropping histories that supported native sagebrush vegetation (NSB), irrigated moldboard plowed crops (IMP), irrigated conservation -chisel- tilled crops (ICT) and irrigated pasture systems (IP). The CO2 emitted as a result of fertilizer production, farm operations and CO2 lost via dissolved carbonate in irrigation water, over a 30 year period, was included. Net organic C in ecosystems decreased in the order IP>ICT>NSB>IMP. In winter, active fungal, bacterial and microbial biomass was greater in IP soils than all other soils. Active bacterial biomass and the amount of DNA extracted from soils was greater in IP soils than all other soils. NSB soils at 0-30 cm (0-5, 5-15 and 15-30 cm inclusive) had greater bacterial diversity in the V1+V2, V1, V3 and V9 regions than all other soils. ICT soils at 0-30 cm (0-15 and 15-30) had greater bacterial diversity in the V1+V2, V1, V3 and V9 regions than IP or IMP soils. IP soils at 0-30 cm had greater bacterial diversity in the V1+V2, V1, V3 and V9 IMP soils. Bacterial diversity and the similarity index declined in the order NSB 0-5 cm > NSB 5-15 > NSB 15-30 > CT 0-15 cm > IMP = ICT 15-30 > IP.Introduction
Land use changes can impact the amount of C stored in the soil by altering C inputs and losses. In forest, grassland and wetland ecosystems, conversion of native vegetation to agricultural cropping has resulted in substantial C transfer to the atmosphere as a result of loss of climax vegetation to the lower equilibrium C concentration in soil. In arid and semi-arid environments plant survival and growth is limited by available water and irrigation is required to increase plant production increasing C input to soils via increased litter and root production.Biodiversity is important because it is the main index of ecosystem health. Within limits, as the biodiversity of an ecosystem increases, the health and stability of the ecosystem increases; conversely, as ecosystems degrade, ecosystem biodiversity decreases. The objective of this research was to determine if land managed as irrigated moldboard plowed crops converted to irrigated conservation tillage or irrigated pasture could not only sequester additional C, but also increase soil microbial biomass, and functional and genetic microbial diversity.
Materials and Methods
Site Description: The study area is located on the Snake River Plain, between 42° 30' 00" and 43° 30' 00" N. and 114° 20' 00" and 116° 30' 00" W. The area is classified as a temperate semi-desert ecosystem. Vegetation throughout the general area was historically dominated by basin big sagebrush (Artemisia tridentata var. tridentata Nutt) and perennial bunch grasses. Experimental Design: The experiment was arranged in a completely randomized design. Soil samples were taken from: 1) three sites supporting native sagebrush vegetation (NSB) located near agricultural land in Southern Idaho (each site supported a basin big sage and a Wyoming big sage vegetation type) ; 2) three sites that were formerly crop land and converted to and maintained as irrigated pasture (IP) for the past 30 years; 3) three sites that were irrigated crop land and have been managed with conservation tillage (ICT) for the past 8 years and 4) three irrigated agricultural crop lands in moldboard plowing systems (IMP).Soil Sampling: We sampled the top 30 cm of soil each season to determine if the amount of C in soil would be affected by vegetation and irrigation. Sampling locations were randomly chosen at each site or field. Separate 10 cm diameter cores were taken and partitioned into 0-5 cm, 5-15 cm, 15-30 and 30-100 cm depths. Roots greater than 1.0 cm diameter were measured separately.
Carbon and Microbial Biomass Analysis: Concentration of organic C in each sample of mineral soil was determined by the Walkley-Black procedure. O organic C was also determined by loss on ignition. Inorganic C was estimated by subtracting organic C from total C. The amount of C per ha-1 of the 0-100 cm of mineral soil was calculated assuming 0.44 g C g-1 organic matter with correction for soil bulk density. Active and total bacteria and fungi numbers and biomass in leachate and surface flow were determined by staining microorganisms and then direct counting.
DNA Extraction: Whole community genomic DNA was extracted from soil samples using slight modifications to the bead beating FastDNA®SPIN Kit for Soil (QBiogene, Vista, CA). The DNA was further purified with a Microcon-100 microconcentrators (Amicon, Beverly, MA) per manufacturer's protocols.
Amplicon Length Heterogeneity-PCR (LH-PCR): Four primer sets were used in the LH-PCR study but only the forward primer was fluorescently labeled and used with the four separate non-fluorescent reverse primers. The primer sequences were (E. coli numbering): for domains V1 and V2: (27F: 5’- 6-FAM-AGA GTT TGA TCM TGG CTC AG- 3’) and 355R (5’- GCT GCC TCC CGT AGG AGT- 3’); domain V1: P1F: (5’ 6-FAM-GCG GCG TGC CTA ATA CTA GC 3’); P1R (5’- TTC CCC ACG CGT TAC TCA CC 3’): domain V3: 338F (5’ HEX-ACT CCT ACG GGA GGC AGC AG 3’); 518R (5’ ATT ACC GCG GCT GCT GG 3’): domain V9: 1005F (5’ NED ATG GCT GTC GTC AGC T 3’) (PE Biosystems, Foster City, CA); EC 1392R (5’ ACG GGC GGT GTG TRC 3’ were used (all other primers, Invitrogen, Rockville, MD). Fluorescent LH-PCR samples were mixed with a 5:1:1 mixture of deionized formamide:blue dextran-EDTA loading dye: GeneScan-500 ROX internal standard (PE Biosystems, Foster City, CA) and separated using 4.25% denaturing polyacrylamide gels (19:1 bis:acrylamide, Bio-Rad, Richmond, CA) on a ABI™ 377 DNA sequencing instrument.
All LH-PCR fingerprint profiles were collected and analyzed by the ABI Prism™ GeneScan® and ABI Prism™ Genotyper® softwares (PE Biosystems, Foster City, CA). Three replicate profiles for each sample were compared to assess the reproducible fragments that could be used for analyses. Descriptive statistics were performed using Microsoft Excel and the averaged relative ratios were used in subsequent analyses. The Shannon diversity index (H), richness (S) and evenness (E) values were calculated.
Statistical Analysis: Carbon data were subjected to a one way vegetation type analysis of variance (ANOVA) for a completely randomized design. Significance of treatment means were determined at P < 0.05 with the Least Square Means test.
DNA Extraction: The amount of DNA extracted from soils declined in the order IP 0-30 cm depth >ICT 0-15 cm depth > IMP 0-30 cm depth >NSB 0-5 cm depth = ICT 15-30 cm depth > NSB 5-15 cm depth = NSB 15-30 cm depth.
Results
Carbon: Soil C as measured by Walkley Black and loss on ignition was higher in the NSB 0-5 cm soil depth than the 5-15, 15-30 and 30-100 cm depths and all other soils. Soil C as measured by Walkley Black and loss on ignition was higher in the 0-30 cm depth increment in the IMP, ICT and IP than the 30-100 cm soil depths. After adjustment for CO2 emissions from farm operations, net C in soils declined in the order IP>ICT>NSB>IMP.Microbial Biomass: In winter, active fungal, bacterial and microbial biomass was greater in IP soils than all other soils. In summer, active fungal biomass was greater in IMP soils. In summer active bacterial, fungal and microbial biomass correlated with soil Walkely Black C in positive curvilinear relationships (r2=0.76, 0.99 & 0.70 respectively). In winter active bacterial, fungal and microbial biomass correlated with soil Walkely Black C in positive curvilinear relationships (r2=0.93, 0.80 & 0.76 respectively).
The amount of microbial DNA extracted from soil correlated with soil Walkely Black C in a positive curvilinear relationship (r2=0.70). In summer, active fungal biomass was greater in IMP soils. The amount of microbial DNA extracted from soil correlated with active bacterial biomass, active fungal biomass and active microbial biomass in positive curvilinear relationships (r2=0.99, 0.89 & 0.94 respectively).
Amplicon Length Heterogeneity: NSB soils at 0-30 cm (0-5, 5-15 and 15-30 cm inclusive) had greater bacterial diversity in the V1+V2, V1, V3 and V9 regions than all other soils. ICT soils at 0-30 cm (0-15 and 15-30) had greater bacterial diversity in the V1+V2, V1, V3 and V9 regions than IP or IMP soils. IP soils at 0-30 cm had greater bacterial diversity in the V1+V2, V1, V3 and V9 IMP soils (Figures 1 and 2). Bacterial diversity and the similarity index declined in the order NSB 0-5 cm > NSB 5-15 > NSB 15-30 > CT 0-15 cm > IMP = ICT 15-30 > IP.
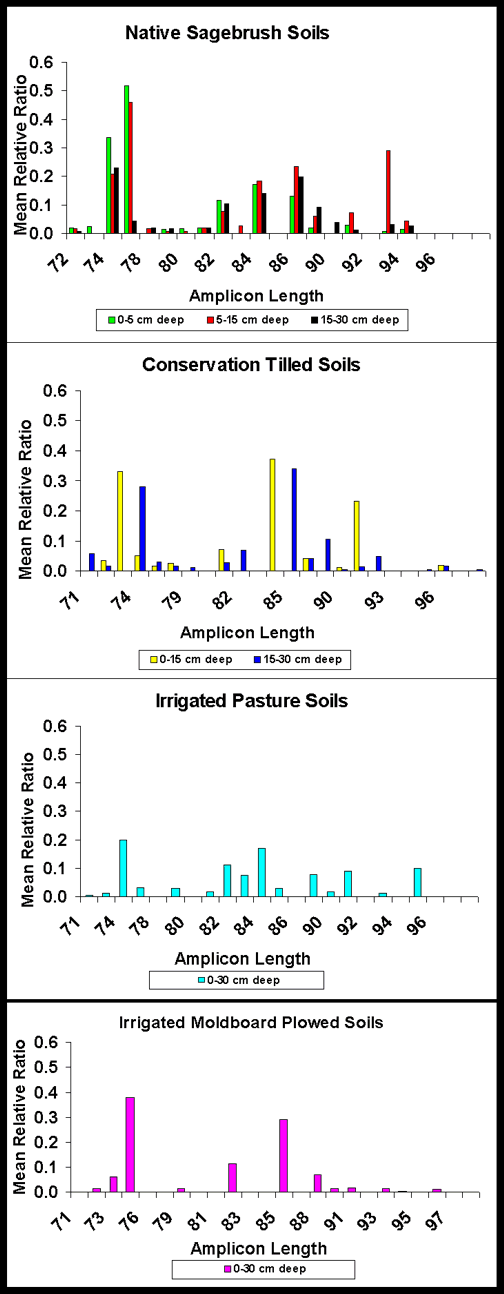 V1 Region Figure 1. Comparison of amplicon lengths in the V1 region of eubacterial 16S rDNA extracted from native sagebrush soils at 0-5, 5-15 and 15-30 cm depths, irrigated conservation tilled soils at 0-15 and 15-30 cm depths, irrigated pasture soils at 0-30 cm depths and irrigated moldboard plowed soils at 0-30 cm depths. |
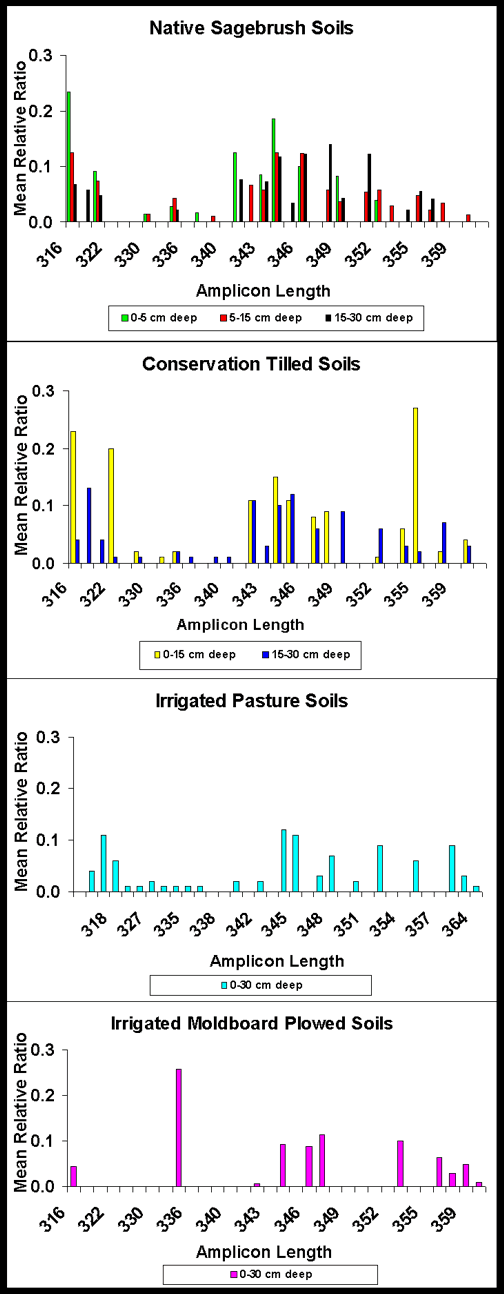 V1 + V2 Region Figure 2. Comparison of amplicon lengths in the V1 + V2 region of eubacterial 16S rDNA extracted from native sagebrush soils at 0-5, 5-15 and 15-30 cm depths, irrigated conservation tilled soils at 0-15 and 15-30 cm depths, irrigated pasture soils at 0-30 cm depths and irrigated moldboard plowed soils at 0-30 cm depths. |
Discussion
As ecosystems mature, they accumulate soil C to a maximum carrying potential, which is controlled by climate, topography, soil type and vegetation. Therefore, at equilibrium, the amount of C added to the soil via vegetation is equal to the amount of C lost through organic matter degradation and other losses and eventually a maximum limit will be reached. Although irrigated agricultural systems can not accrue C indefinitely, with improved management they can potentially remove substantial amounts of C from the atmosphere for the next 30-50 years.Irrigated agriculture increased active bacterial and fungal biomass and functional diversity, but decreased bacterial diversity in these soils. We may be observing that few species of bacteria are performing the same biological functions in irrigated soils compared to native sagebrush soils. Since biodiversity is an index of ecosystem health and as the biodiversity of an ecosystem increases, the health and stability of the soil ecosystem should increase. Irrigated agriculture may not improve bacterial diversity in these soils.
Given that our society must produce food, conservation tillage seems to result in greater bacterial functional and genetic biodiversity than conventional (moldboard plowed) tillage. When native forest or grassland is converted to agriculture large amount of C are transferred from soils to the atmosphere with the associated loss of diversity of plants and soil microorganisms. Since, irrigated pasture and conservation tilled systems sequester more C than native sagebrush an argument can be made to expand irrigated conservation tilled agriculture and pastures recognizing the slight loss in bacterial genetic diversity in the soil ecosystem for the foreseeable future.
Click to view a bigger image of this poster.
Back to PAM Research Project Page
*Disclaimer*
Posters, by their nature, are preliminary findings. These data and their interpretation may be refined or changed before formal publication. We encourage the user to benefit from their early insight into our most recent research, but not to regard them as the "final word" on these findings.