| Simon: Posters: 3 |

|
American Society for Horticultural Science 98th Annual Conference & Exhibition - July 22 - 25, 2001
Genetic Diversity in Garlic (Allium sativum L.) as Assessed by AFLPs and IsozymesMeryem Ipek1 & Philipp Simon1,2 |
ABSTRACT
Garlic (Allium sativum L.), an asexually propagated crop, displays great morphological diversity. The genetic diversity of 45 garlic clones and three A. longicuspis clones, the proposed progenitor of A. sativum, was evaluated using AFLP. Three primer combinations of EcoR I + 4 and Mse I + 3 (where + 4 and + 3 are the number of selective bases used) generated a total of 183 polymorphic fragments. The Jaccard's similarity matrix was used for UPGMA cluster analysis. Although similarities between the clusters were low (? 0.30), some clones within the clusters were very similar (<0.95) with AFLP analysis. Furthermore, 16 clones represented only six different types, within which they shared 100% polymorphic AFLPs. These may, therefore, be duplicates. In agreement with the results of other investigators, A. longicuspis and A. sativum clones were clustered together with no clear separation, suggesting these species are not genetically distinct. A subset of 48 clones has been analyzed with polymorphic isozyme systems. Cluster analysis of AFLP has been compared with the results of isozyme analysis.
INTRODUCTION
Cultivated garlic (Allium sativum L.), known at least 5000 years (Hahn, 1996) and its proposed ancestor, Allium longicuspis Regl. originated in Central Asia (Vavilov, 1935). Although garlic is an asexually propagated crop, it displays great morphological diversity in bulb, and leaf size, color and shape, scape presence and height, and flower color, fertility, and bulbil (topset) development (Pooler and Simon, 1993).
Morphological characters may differ under varying environmental conditions. This can complicate classification of garlic. Isozyme and RAPD markers have been utilized to classify and categorize genetic diversity in garlic (Al-Zahim et al., 1997; Bradley et al., 1996; Lallemand, 1997; Maass and Klaas, 1995; Pooler and Simon, 1993). In this study, we have utilized the AFLP technique to evaluate genetic variation among the diverse garlic clones including its proposed ancestor, A. longicuspis, collected from different parts of the world. In addition, we have performed isozyme assays and compared both systems for the assessment of genetic diversity in garlic.
MATERIALS & METHODS
Young and healthy leaves were sampled from one to three plants from each clone, freeze-dried, and DNA was extracted according to previously described 1•-CTAB method (Murray and Thompson 1980).
AFLP protocols were followed according to the manufacturer's procedure with minor modification using the AFLP Kit number I (Gibco-BRL Life Technologies Inc.) for plant species with large genomes. Half of the recommended reaction volumes were used except pre-selective amplification that was performed at 1/5 the recommended volume. All polymorphic bands were identified and numbered in order, starting with the largest polymorphic fragment. Only unambiguously polymorphic bands were scored as present or absent. Similarity matrices were generated according to the coefficient of Jaccard (Sneath and Sokal, 1973). This similarity matrix was used to perform a cluster analysis using the un-weighted pair-group method with arithmetic averaging (UPGMA) (Sokal and Michener, 1958) with the software NTSYS-PC (Numerical Taxonomy and Multivariate Analysis System version 1.80) (Rohlf, 1993).
Isozyme analysis was carried out according to the methods described by Pooler and Simon (1993). Bands were scored directly from the PAGE gels as present or absent. A similarity matrix prepared using Simple Matching (SM) coefficient was used for UPGMA cluster analysis.
RESULTS & DISCUSSION
 |
 |
| E+3-M+3 | E+4-M+3 | |
|---|---|---|
 |
 |
Preliminary evaluations with EcoR I and Mse I selective amplification primers having three base selective nucleotide extensions yielded more than 250 bands and data analysis was ambiguous. Since garlic has a large genome size of over 3X1010 bp (Ranjekar et al., 1978), four base nucleotide extended EcoR I selective amplification primers were used to reduce the number of amplified bands to between 130-150 bands, which could be scored accurately. |
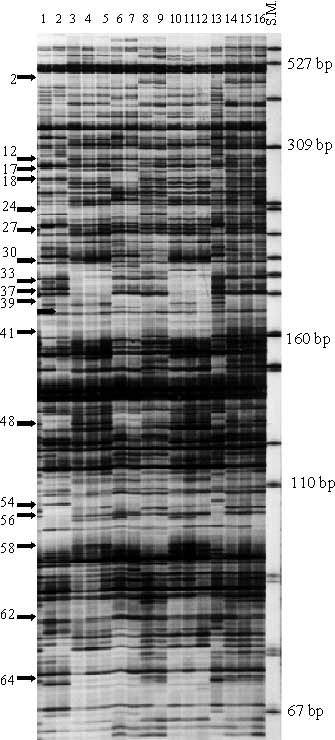 |
Three combinations of EcoR I + 4 and Mse I + 3 selective amplification primers (EcoR I + ACGA / Mse I + CAT, EcoR I + ACGG / Mse I + CTC, and EcoR I + ACGG / Mse I + CAT), generated 64, 63 and 56 AFLPs respectively for a total number of 183 polymorphic AFLPs among 45 garlic and three A. longicuspis clones. Scored AFLPs ranged from 67 bp to 457 bp in size. |
| DIA | Esterase | PGM | G6PDH |
|---|---|---|---|
 |
 |
 |
 |
For isozyme analysis, consistently resolving 8 polymorphic band were scored, 3 for 6PDH, 2 each for EST and PGM, and 1 for DIA.

The UPGMA cluster analysis of isozyme data placed 39 garlic clones into 6 clusters. Similarities among the clusters ranged from 0.24 to 0.86 according to the simple matching coefficient. Within each cluster, clones shared 100% of isozyme markers.
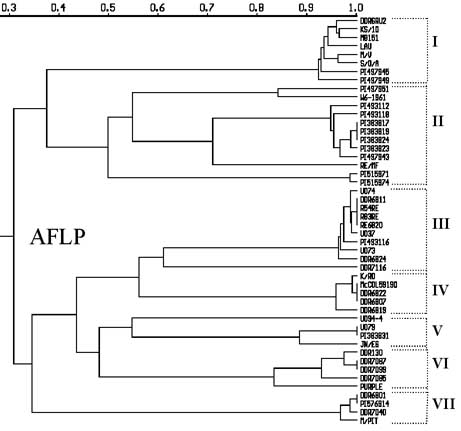
The UPGMA dendrogram constructed using the Jaccard's similarity matrix based on the shared fragments of 183 polymorphic AFLPs . Sixteen clones were represented in six clusters by sharing 100% of 183 AFLPs within each group: 1) PIs 383817, 383819, and 383824; 2) DDR6811, RE/6820, R54RE, and R83RE; 3) McCOL59190, DDR6822, and DDR6807; 4) U079 and PI 3838331: 5) DDR7087 and DDR7099; and 6) DDR6801 and PI 576914. These likely are duplicates. In addition, the AFLP analysis of this study found 36 out of 48 clones to be grouped into nine clusters where within each cluster a similarity of 0.95 or greater was noted based on dendrogram (e.g. DDRGRU2, M8151, and KS/10).
CONCLUSIONS
- EcoR I + 4 andMse I + 3 primers combinations can be used for genetic diversity studies in garlic with AFLP.
- Despite the clonally propagated nature of garlic, we found that genetic diversity revealed by AFLP analysis was high between groups.
- The UPGMA cluster analyses of AFLP and isozyme data were comparable. Clones sharing more than 80% of the AFLPs were clustered together in the same group by isozyme markers.
- Although isozyme analysis gave similar grouping with AFLP analysis, AFLP with much large number of markers gave a better approximation to true variation among clones.
- In the UPGMA AFLP dendrogram, all non-bolting garlic clones were grouped in cluster II. Cluster I contains bolting but early senescing garlic clones, and the remaining clusters contained other bolting garlic clones.
- The AFLP technique is a useful tool for analysis of garlic genetic diversity along with morphological characters, isozymes, and RAPDs.
References
Al-Zahim, M., H. J. Newbury, and B. V. Ford-Lloyd. 1997. Classification of genetic variation in garlic (Allium sativum L.) revealed by RAPD. HortScience 32:1102-1104.
Bradley, K. F., M. A. Rieger, and G. G. Collins. 1996. Classification of Australian garlic cultivars by DNA fingerprinting. Aust. J. Exp. Agric. 36:613-618.
Hahn, G. 1996. History, folk medicine and legendary uses of garlic, p. 1-34. In: H. P. Koch and L. D. Lawson (eds.) Garlic, the science and therapeutic application of Allium sativum L. and related species (2nd edition), Williams and Wilkins, Baltimore, Maryland.
Lallemand, J., C. M. Messian, F. Briad, and T. Etoh. 1997. Delimitation of varietal groups in garlic (Allium sativum L.) by morphological, physiological and biochemical characters, p. 123-132. In: J. L. Burba and C. R. Galmarini (eds.) Proc. I Int. Symp. Edible Alliaceae, Acta Hort. 433.
Maass, H. I. and M. Klaas. 1995. Infraspecific differentiation of garlic (Allium sativum L.) by isozyme and RAPD markers. Theor. Appl. Genet. 91:89-97.
Murray, J. M. and W. Thompson. 1980. Rapid isolation of high-molecular weight plant DNA. Nucleic Acids Res. 8:4321-4325.
Pooler, M. R. and P. W. Simon. 1993. Characterization and classification of isozyme and morphological variation in a diverse collection of garlic clones. Euphytica 68:121-130.
Ranjekar, P. K., D. Pallotta, and J. G. Lafontaine. 1978. Analysis of plant genomes. V. Comparative study of molecular properties of DNAs of seven Allium species. Biochem. Genet. 16:957-970.
Rohlf, F. J. 1993. NTSYS-pc numerical taxanomy and multivariate analysis system. Version 1.8. Exeter Publ., Setauket, NY.
Sneath, P. H. A. and R. R. Sokal. 1973. Numerical taxonomy. Freeman, San Francisco.
Sokal, R. R. and C. D. Michener. 1958. A statistical method for evaluating systematic relationships. Univ. Kansas Sci. Bull. 38:1409-1438.
Vavilov, N. I. and V. F. Dorofeev. 1935. The phyto-geographical basis for plant breeding [first published in Russian, translated by D. Love (1992)]. In: V. F. Dorofeyev (eds.) Origin and geography of cultivated plants (English edn.). University Press, Cambridge.
