| Soybean Quality Improvement Laboratory |

|
Problem
Soybeans are an important crop because of the quantity and quality of protein and oil in the seeds. Seed storage protein and fatty acids accumulate during seed development, and this process depends upon precise orchestration of gene transcription - both genetic and environmental factors influence the eventual composition of seeds. To understand these factors we study soybean seed development. The goal of this work is to make soybean plants with a more nutritious balance of proteins in the seed, and oils that are either healthier or better suited for industrial applications, and to understand how events that occur during seed development impact yield.
Current Projects in the laboratory include:
Gene expression during soybean seed development
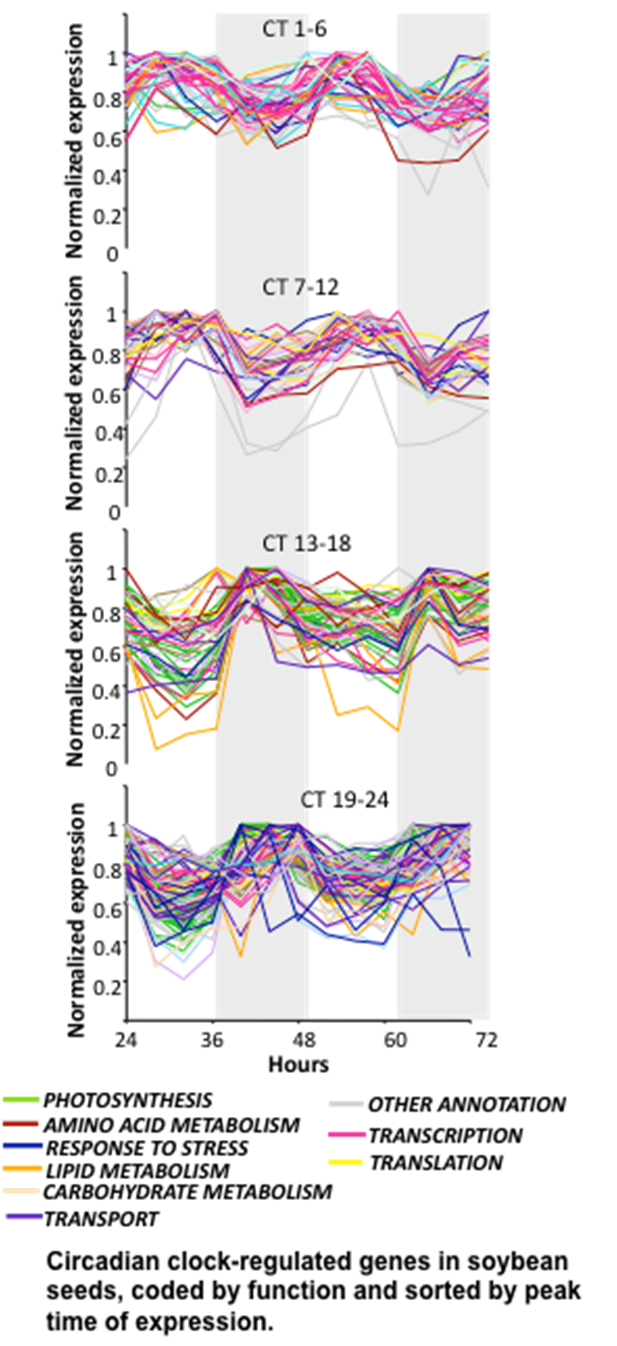
In order to understand how the expression of genes relevant to seed composition and yield is controlled, the laboratory studies networks of gene regulation in developing soybean seeds. Recent work has focused on the control of seed gene expression by diurnal cycles or the plant circadian clock. Many of the biological and physiological processes involved in protein biosynthesis, plant secondary metabolism, and carbohydrate metabolism are known to be controlled by the circadian clock or influenced by diurnal cycles. However, it was not known to what extent these factors influence gene expression during seed development. We used microarray expression profiling to identify genes that are regulated by the circadian clock in developing soybean seeds. Over 400 genes in developing seeds show evidence of circadian regulation, and these genes encode proteins that are predicted to be involved in many different metabolic processes in the seeds. Interesting differences in gene expression were observed between leaves and seeds of the same plants in circadian clock controlled gene expression and diurnal cycles. A number of transcriptional regulators that are controlled by the circadian clock in soybean seeds were identified, and are currently being characterized. In addition, promoter motifs were found to be common to subsets of co-regulated genes, which represent potential targets of transcription factor binding.
We are also using several parallel approaches to characterize the expression and function of candidate genes in soybean that are believed to be important for seed development and composition based on their roles in seed development in other plant species.
Soybean TILLInG population
 The soybean TILLInG (Targeting Induced Local Lesions In Genomes) population is a resource of induced mutant soybean plants that are being screened for genetic variation in useful traits. Forward genetic screening of this population has identified genetic loci involved in determining seed responsiveness to the plant hormone abscisic acid, and loci required for the normal fatty acid composition of soybeans. In addition, we are testing ways to use the population for reverse genetic screening to identify mutations in genes that may be of interest to other soybean researchers.
The soybean TILLInG (Targeting Induced Local Lesions In Genomes) population is a resource of induced mutant soybean plants that are being screened for genetic variation in useful traits. Forward genetic screening of this population has identified genetic loci involved in determining seed responsiveness to the plant hormone abscisic acid, and loci required for the normal fatty acid composition of soybeans. In addition, we are testing ways to use the population for reverse genetic screening to identify mutations in genes that may be of interest to other soybean researchers.
