| Use of the Hsp90 gene for nematode molecular phylogenetics |

|
 The use of multiple molecular markers strengthens both diagnosis and the systematics of nematodes. Traditionally, ribosomal and mitochondrial genes have been the most popular genes for this purpose, largely because they are present in multiple copies and are easy to amplify. A few other genes, including the RNA polymerase II large subunit, have been applied for nematode systematics, but generally, few have caught on.
The use of multiple molecular markers strengthens both diagnosis and the systematics of nematodes. Traditionally, ribosomal and mitochondrial genes have been the most popular genes for this purpose, largely because they are present in multiple copies and are easy to amplify. A few other genes, including the RNA polymerase II large subunit, have been applied for nematode systematics, but generally, few have caught on.
We have introduced the gene for Hsp90 as a new marker useful for molecular systematics of nematodes. Hsp90 has been used for phylogenetic analysis in a variety of other organisms and has a good balance of sequence conservation and diversity. It is part of a heat shock protein gene family, but has distinct domain structure and signature sequences that allow specific amplification of Hsp90 and not other family members. Therefore, some of the potential problems that can arise from use of paralogous genes (e.g., Hsp70) are not an issue with Hsp90.
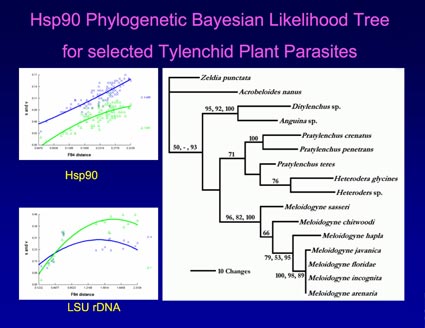 We have performed an in-depth analysis of Hsp90 and compared trees based upon Hsp90 alignments with those based upon 18S and 28S ribosomal gene alignments. We have found Hsp90 cDNA sequences to be especially useful for constructing genus and species level phylogenies. Amino acid alignments have sufficient signal for broader-scale taxonomic comparisons. We have built a growing database of partial Hsp90 sequences (focusing on a ~2 kb region from the center of the gene), which have been deposited in GenBank and are searchable at www.ncbi.nlm.nih.gov.
We have performed an in-depth analysis of Hsp90 and compared trees based upon Hsp90 alignments with those based upon 18S and 28S ribosomal gene alignments. We have found Hsp90 cDNA sequences to be especially useful for constructing genus and species level phylogenies. Amino acid alignments have sufficient signal for broader-scale taxonomic comparisons. We have built a growing database of partial Hsp90 sequences (focusing on a ~2 kb region from the center of the gene), which have been deposited in GenBank and are searchable at www.ncbi.nlm.nih.gov.
Skantar, A. M. and Carta, L. K. Phylogenetic evaluation of nucleotide and protein sequences from the heat shock protein 90 gene of selected nematodes. J. Nematol. 36 (4): 466-480. 2005
