Research Microbiologist
Research Objectives
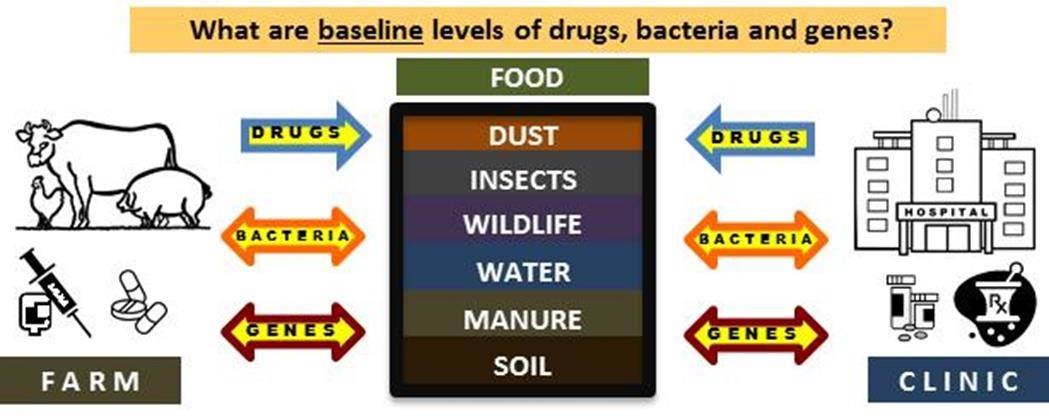
- Tracking Agricultural Antibiotic Resistance (AgAR). Antibiotic resistance is an urgent global health crisis. Agricultural producers and researchers need tools to help them measure resistant bacteria and resistance genes in soil, water, air, and the environment. My research objectives in this area are to develop and validate reliable, high-quality methods to find and count antibiotic resistant bacteria and their genes in farms and on fields, and use these tools to understand the ecology of antibiotic resistance in agroecosystems. The focus of these projects is on resistance classes that are microbiologically important based on carriage of likely pathogens, clinically important based on the kinds of drugs used to treat infections in food animals and humans, and ecologically important based on regional soil and water systems.
- Profile Naturally Occurring Antibiotic Resistance. Antibiotic resistant bacteria and genes occur naturally in soils across the globe. From the perspective of human health, even the naturally occurring antibiotic resistance is a potential threat. My research objective is to improve the usefulness of agricultural antibiotic resistance data so that it is possible to effectively identify specific types of resistance that can be impacted by agricultural best management practices.
- Fighting Foodborne Pathogens. Recent outbreaks highlight the need to know how long specific E. coli strains survive in the environment. My research objective in this area is to provide information and tools to assist public health professionals and agricultural producers to they can control illness and deaths associated with these organisms. Currently, I am conducting studies to measure how long non-O157 Shiga-toxigenic E. coli survive in manure-impacted runoff.
- Uncover Links Between Microbial Community Structure and Soil Organic Carbon Storage. The organic carbon stored in soils is important in crop production, soil health, and the cycling of carbon through the ecosystem. Microbes play a critical role in these processes. Agronomists at the Lincoln ARS have evidence of deep soil carbon storage in long-term plots growing crops, and I am working with the team to uncover which microbes might be responsible for these deep carbon stores in the soil.
|
|
|
1. Tracking Agricultural Antibiotic Resistance (AgAR). Regardless of whether resistance occurs naturally or is the result of human activities, we need to better determine how to mitigate potential health risks from environmental bacteria and genes. This includes gathering information on how they move within food production systems, what conditions encourages them to die or persist in soil, air and water, and how they move between urban and rural environments. Image from Durso and Cook, 2014. Impacts of antibiotic use in agriculture: what are the benefits and risks? Curr Opinion Microbiol 19:37-44. |
|
|
|
2. Profile Naturally Occurring Antibiotic Resistance. Antibiotic resistant bacteria and their genes are ubiquitous in the environment. This often goes unnoticed, since most environmental bacteria do not make people sick. In order to determine the impacts that specific farming or human waste management practices have on environmental resistance, we need to know what kinds and amounts of resistance occurs naturally. Studies performed at Nine-Mile Prairie in southeast Nebraska, pictured above, help us know what kinds and amounts of antibiotic resistant bacteria and antibiotic resistance genes occur naturally in Nebraskan soils. Since over 90% of the land in Nebraska is devoted to agricultural production, these native prairie sites give us an important insight into how specific farming practices have impacted soils over time. |
|
|
|
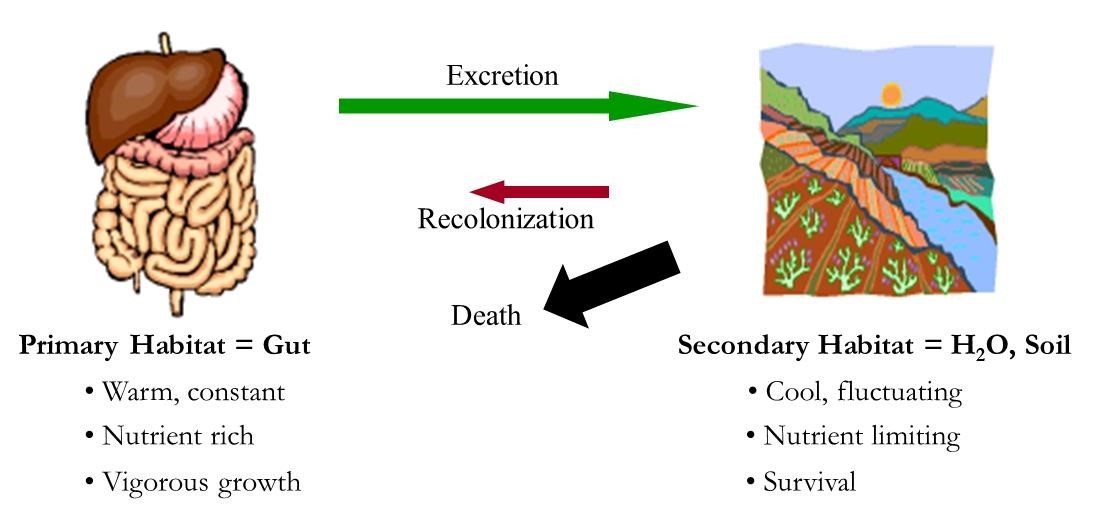
3. Fighting Foodborne Pathogens. Foodborne pathogens that come from farm animals (zoonotic pathogens) live primarily in two main habitats. They usually grow in the lower intestines of warm blooded animals (including humans and farm animals). Once they are excreted from the animal via feces, they must survive or grow in water, soil, and sediment until they find a new host, or they die. I collect information on how well the pathogens survive in the environment, and how different agronomic practices like buffer strips or manure incorporation influence their survival. I am also collecting information on how long specific serotypes of Shiga-toxigenic E. coli (STEC) survive in water and soil. My lab has expertise in finding and isolating STEC from environmental sources, and I work with human public-health labs to help them isolate STEC in outbreaks associated with agricultural events such as fairs and ag-expos. |
|
|
|
4. Uncover Links Between Microbial Community Structure and Soil Organic Carbon Storage. ARS Scientists and technicians collect deep soil cores from long-term (30-+ year) switchgrass plots. There is intense interest in the agricultural and biogeochemical communities for high-resolution microbial community data from the soils that support cropping systems and are linked to long-term management data. This project pairs high resolution taxonomic and functional information on bacterial and fungal communities with our unit’s existing comprehensive collection of soil physical and chemical data. Additionally, decades of information on agronomic and management outcomes is available for these sample plots, providing valuable phenotyping and metadata that can be leveraged to provide a deep and thorough understanding of the role of microbes in C and N cycling. There is interest in these outcomes from both basic and applied scientists. |