| Asis Khan |

|
|
Asis Khan, Ph.D. Animal Parasitic Diseases Laboratory |
 |
Welcome
In contemplating a vision for the future of genetics and genomics research in the post-genomic era, it is important to integrate genetics and genomics to improve health by controlling the emergence and re-emergence of infectious diseases. At the interface between genetics, genomics, and infectious disease, our research focuses on understanding how pathogen evolution influences host, pathogen, and microbiome interactions. Specifically, it is important to reiterate that we are interested in understanding the molecular mechanisms of pathogenesis by identifying genetic determinants of pathogens responsible for host-specificity and host immune responses using evolution-guided approaches, which include population genetics, comparative genomics, and genome-wide association test. To accomplish these long-term goals, our lab utilizes unicellular enteric pathogens including Cryptosporidium, Giardia, and Cyclospora to systematically evaluate the epidemiology, population genetics, genetic determinants of host specificity, and the interplay between host, intestinal microbiota, and the pathogen that affects disease resolution, morbidity, or mortality. Understanding the nature of these complex interactions, particularly in the context of parasite transmission (zoonotic versus anthroponotic), host specificity and disease severity will significantly enhance our ability to effectively combat the devastating enteric pathogens.
Research
A billion cases of illness and millions of deaths take place every year globally due to zoonotic diseases. Strikingly, many foodborne outbreaks are linked to zoonotic pathogens which are urging government intervention and public vigilance against the risks of catching foodborne diseases. Most of these foodborne diseases are associated with diarrheal diseases which are the second leading causes of morbidity and mortality in children less than 5 years of age worldwide and are responsible for ~760,000 child fatalities every year. The most common protozoan agents responsible for persistent diarrhea are Cryptosporidium, Cyclospora, and Giardia; of these, according to a recent Global Enteric Multicenter Study (GEMS), Cryptosporidium is the 2nd leading cause of severe diarrhea and mortality after rotavirus in young children in Africa and southern Asia. In the absence of vaccines, prevention of these parasitic diseases relies solely on chemotherapy, which is far from perfect and limited by adverse drug effects. Thus, there is an urgent need for new drugs/vaccines and furthermore, new genetic technologies to identify potential drug/vaccine targets to combat these parasitic infections.
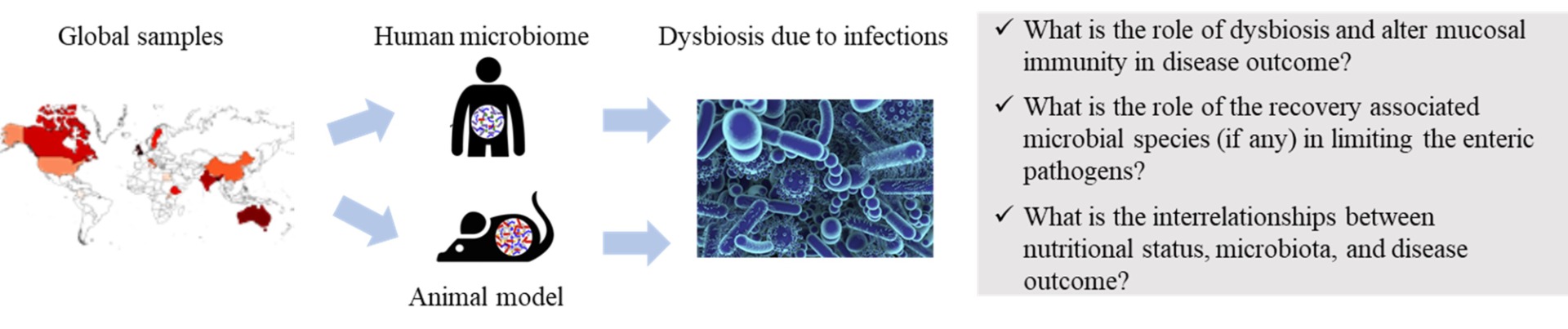
Epidemiology and comparative genomics: In the advent of next-generation sequencing (NGS), comparative genomics of pathogens is the mainstay of new drug and vaccine development. However, NGS based whole genome sequencing (WGS) of medically important eukaryotic parasites is still hindered by some basic obstacles including a) in vitro propagation techniques of these parasites are not well developed to produce sufficient parasite material for WGS, b) no animal model exists to expand protozoan parasites in animals, c) current WGS efforts involve an immunomagnetic separation (IMS) step to enrich parasite oocysts, which must be collected fresh from clinical/field samples without freezing, as the freeze-thaw procedure ruptures oocyst membranes making the IMS technique intangible, and d) WGS is typically limited to symptomatic samples that possess relatively high parasite burdens (≥103 oocysts per gram, OPG). Strikingly, asymptomatic carriage of these parasites in indigenous populations is very common (~40 to 70%) and parasite oocyst numbers in asymptomatic patients are limited, but not below the lower limit of detection for target enrichment sequencing (TES). To overcome these obstacles, we are developing highly specific and sensitive TES for whole genome amplification of protozoan parasites directly from fecal samples, which represents a novel step in adapting TES methodology for whole genome amplification of pathogens followed by next-generation sequencing and population genomic assay in complex biological samples containing a low pathogen/host nucleic acid ratio. We seek to combine comparative genomics and genome wide association tests by utilizing the whole genome sequences directly from the fecal samples to identify and validate candidate determinates for differential diseases outcomes (symptomatic vs. asymptomatic) and host specificity using reverse genetics and CRISPR/Cas9-based gene knockout and complementation experiments in mice and human intestinal organoid cultures. By identifying candidate determinates and therapeutic targets for anti-parasitic drugs, our study will pursue fundamental insights concerning eukaryotic microbiology and contribute new approaches to mitigating the harms imposed by these parasites, especially to children who suffer considerable illness and death from the parasitic enteric diseases.

Interrelationship between nutritional status, microbiota, and disease outcome due to parasitic infection: Worldwide, ~200 million children under the age of 5 are suffering from stunted growth, and it has been well documented that infection with enteric pathogens (including Cryptosporidium, Cyclospora, and Giardia) and malnutrition are intricately connected to this phenomenon. One puzzling observation is that in the endemic regions, infections with enteric parasites can often be asymptomatic which we believe to be due to specific parasite genetic factors, gut microbiome, and/or host immune and nutritional status. Evidence now exits for bidirectional communication between the three key factors: nutritional status, immune system, and commensal microflora, which can have a profound influence on the disease outcome due to infection of enteric pathogens. To better understand the association between parasitic interactions, malnutrition, and the gut microbiome, we are interested to determine the association of the members of the gut microbiota with severe human cryptosporidiosis or giardiasis in malnourished compared to healthy children. This proposal will provide an understanding of the role of the gut microbiota that precedes severe diarrhea due to enteric pathogens in malnourished compared to healthy children.
Members
Asis Khan
Education:
B.Sc., Calcutta University, India
Ph.D., Jadavpur University, India
Postdoctoral Fellow, Washington University School of Medicine, St. Louis, USA
Email: asis.khan@usda.gov
CV (pdf)
Post-Doctoral Fellows
Coming Soon
We are currently seeking post-doctoral candidates. Interested applicants can send an email to asis.khan@usda.gov with a CV and summary of previous research experience and future interests.
Position #1: Research Opportunity in Host Parasite Interaction (through ORISE and USDA/ARS)
Position #2: Postdoctoral research associate in Cryptosporidium Genomics (through University of Georgia
Publications
For a complete listing of publications see PubMed Search and Google Scholar Page
Selected Publications:
• Global selective sweep of a highly inbred genome of the cattle parasite Neospora caninum.
Khan A, Fujita AW, Randle N, Regidor-Cerrillo J, Shaik JS, Shen K, Oler AJ, Quinones M, Latham SM, Akanmori DB, Cleveland S, Ryan U, Slapeta G, Schares G, Ortega-Mora LM, Dubey JP, Wastling JM, Grigg ME. Proc Natl Acad Sci USA. 2019; 116 (45): 22764-22773.
PMID:31636194
• Neosporosis: an animal disease.
Khan A#, Shaik JS, Sikorski P, Dubey JP, and Grigg ME. Engineering. 2019. doi: https://doi.org/10.1016/j.eng.2019.02.010 (published to Special Issue on Animal Disease Research, Engineering, Elsevier; invited review)
# Corresponding author
• Genomics and molecular epidemiology of Cryptosporidium species.
Khan A#, Shaik JS, and Grigg ME. Acta Tropica. 2018; 184:1-14.
# Corresponding author
PMID: 29111140
• Toxoplasma gondii: Laboratory maintenance and growth.
Khan A#, and Grigg ME. Curr Protoc Microbiol. 2016; 44:20C.1.1-20C.1.17.
# Corresponding author
PMID: 28166387
• Local admixture of amplified and diversified secreted pathogenesis determinants shapes mosaic Toxoplasma gondii genomes.
Lorenzi H*, Khan A*, Behnke MS*, Namasivayam S, Swapna LS, Hadjithomas M, Karamycheva S, Pinney D, Brunk B, Ajioka JW, Ajzenberg D, Boothroyd JC, Boyle JP, Darde MS, Diaz-Miranda, Dubey JP, Fritz HM, Gennari SM, Gregory DB, Kim K, Saeij J, Su C, White MW, Zhu XQ, Howe DK, Rosenthal BM, Grigg ME, Parkinson J, Liu L, Kissinger JC, Roos DS, and Sibley LD. Nat Commun. 2016 Jan 7;7:10147.
*Co-first authors
PMID: 26738725
• Rhoptry proteins ROP5 and ROP18 are major murine virulence factors in genetically divergent South American strains of Toxoplasma gondii.
Behnke MS*, Khan A*, Lauron EJ, Jimah JR, Wang Q, Tolia NH, and Sibley LD. PLoS Genet. 2015; 11(8):e1005434. eCollection 2015 Aug.
*Co-first authors
PMID: 26291965
• Globally diverse Toxoplasma gondii isolates comprise six major clades originating from a small number of distinct ancestral lineages.
Su C*, Khan A*, Zhou P, Majumdar D, Ajzenberg D, Darde ML, Zhu XQ, Ajioka JW, Rosenthal BM, Dubey JP, and Sibley LD. Proc Natl Acad Sci USA. 2012; 109(15):5844-5849.
*Co-first authors
PMID: 22431627
• Virulence differences in Toxoplasma mediated by amplification of polymorphic pseudokinases.
Behnke M, Khan A, Wootton J, Dubey JP, and Sibley LD. Proc Natl Acad Sci USA. 2011; 108(23):9631-9636.
PMID: 21586633
• Genetic analyses of atypical Toxoplasma gondii strains reveals a forth clonal lineage in North America.
Khan A, Dubey JP, Su C, Ajioka JW and Rosenthal BM, and Sibley LD. Int J Parasitol. 2011; 41(6):645-655.
Accepted for cover page.
PMID: 21320505
• Selection at a single locus leads to widespread expansion of Toxoplasma gondii lineages that are virulent in mice.
Khan A*, Taylor S*, Ajioka JW, Rosenthal BM, and Sibley LD. PLoS Genet. 2009 Mar;5(3):e1000404. Epub 2009 Mar 6.
*Co-first authors.
Reviewed by Faculty of 1000 Biology.
PMID: 19266027
• Recent transcontinental sweep of Toxoplasma gondii driven by a single monomorphic chromosome.
Khan A, Fux B, Su C, Dubey JP, Darde ML, Ajioka JW, Rosenthal BM and Sibley LD. Proc Natl Acad Sci USA, 2007; 104(37):14872-14877.
Featured in: i) Sweeping through Toxoplasma, Editor’s Choice; Science. 2007; 317:1651-1652.
ii) Toxoplasma’s shared heritage, In this issue Proc Natl Acad Sci USA. 2007; 104(37):14872.
PMID: 17804804
• Genetic divergence of Toxoplasma gondii strains associated with ocular toxoplasmosis, Brazil.
Khan A, Jordan C, Muccioli C, Vallochi AL, Rizzo LV, Bellfort R Jr, Vitor RW, Silveira C, and Sibley LD. Emerg. Infect Dis. 2006; 12(6): 942-949.
PMID: 16707050
• Common inheritance of chromosome Ia associated with clonal expansion of Toxoplasma gondii.
Khan A*, Böhme U*, Kelly KA*, Adlem E, Brooks K, Simmonds M, Mungall K, Quail MA, Arrowsmith C, Chillingworth T, Churcher C, Harris D, Collins M, Fosker N, Fraser A, Hance Z, Jagels K, Moule S, Murphy L, O'Neil S, Rajandream MA, Saunders D, Seeger K, Whitehead S, Mayr T, Xuan X, Watanabe J, Suzuki Y, Wakaguri H, Sugano S, Sugimoto C, Paulsen I, Mackey AJ, Roos DS, Hall N, Berriman M, Barrell B, Sibley LD, and Ajioka JW. Genome Research. 2006; 16(9): 1119-1125.
*Co-first authors
PMID: 16902086
• Genotyping of Toxoplasma gondii strains from immunocompromised patients reveals high prevalence of type I strains.
Khan A, Su C, German M, Storch GA, Clifford DB, and Sibley LD. J. Clin. Mirobiol. 2005; 43(12): 5881-5887.
PMID: 16333071
• Composite genome map and recombination parameters derived from three archetypal lineages of Toxoplasma gondii.
Khan A, Taylor S, Su C, Mackey AJ, Boyle J, Cole R, Glover D, Tang K, Paulsen IT, Berriman M, Boothroyd JC, Pfefferkorn ER, Dubey JP, Ajioka JW, Roos DS, Wootton JC, and Sibley LD. Nucl. Acids Res. 2005; 33(9): 2980-2992.
Accepted for cover page
PMID: 15911631
• Shiga toxin producing Escherichia coli infection: current progress & future challenges.
Khan A, Datta S, Das SC, Ramamurthy T, Khanam J, Takeda Y, Bhattacharya SK, and Nair GB. Indian J Med Res. 2003; 18:1-24 Review.
PMID: 14748461
• Environmental isolates of Citrobacter braakii that agglutinate with Escherichia coil O157 antiserum but do not possess the genes responsible for the biosynthesis of O157 somatic antigen.
Khan A, Nandi RK, Das SC, Ramamurthy T, Khanam J, Shimizu T, Yamasaki S, Bhattacharya SK, Chicumpa W, Takeda Y, and Nair GB. Epidemiol. Infect. 2003; 130(2): 179-186.
PMID: 12729185
• Antibiotic resistance, virulence gene, and molecular profiles of Shiga toxin-producing Escherichia coil isolates from diverse sources in Calcutta, India.
Khan A, Das SC, Ramamurthy T, Sikdar A, Khanam J, Yamasaki S, Takeda Y, and Nair GB. J Clin Microbiol. 2002; 40(6): 2009-2015.
PMID: 12037056
• Prevalence and genetic profiling of virulence determinants of non-O157 Shiga Toxin-Producing Escherichia coil isolated from cattle, beef and humans, Calcutta, India.
Khan A, Yamasaki S, Sato T, Ramamurthy T, Pal A, Datta S, Chowdhury NR, Das SC, Sikdar A, Tsukamoto T, Bhattacharya SK, Takeda Y, and Nair GB. Emerg. Infect Dis. 2002; 8(1): 54-62.
PMID: 11749749
Book Chapters:
• Molecular epidemiology and population structure of Toxoplasma gondii.
Darde ML, Mercier A, Su C, Khan A, and Grigg ME. Toxoplasma Gondii, 3rd Edition, The model apicomplexan: perspectives and Methods. Edited by Professor Louis M. Weiss and Professor Kami Kim. Academic Press.
ISBN: 978-0-12-396481-6
• Genetics and genome organization of Toxoplasma gondii.
Khan A, Taylor S, Su C, Sibley LD, Paulsen I, and Ajioka JW. The Biology of Toxoplasma gondii. Toxoplasma: Molecular and Cellular Biology. Edited by James W. Ajioka and Dominique Soldati. Horizon Bioscience.
ISBN 13:978-1-904933-34-2
• Pathogenicity and virulence in Toxoplasma gondii. Toxoplasma: Molecular and Cellular Biology.
Taylor S, Khan A, Su C, and Sibley LD. Edited by James W. Ajioka and Dominique Soldati. Horizon Bioscience.
ISBN 13:978-1-904933-34-2
Contact
|
Laboratory:
|
 |
