| Spiroplasma Kunkeliigenome Sequencing Project (Dr. Davis) |

|
Spiroplasma kunkelii
Genome Sequencing Project
This is the Spiroplasma kunkelii Genome Sequencing Project Web Site, a World Wide Web (WWW) site of the Molecular Plant Pathology Laboratory (MPPL) at USDA-ARS in Beltsville, Maryland. This Web Page provides genomic and other information about S. kunkelii, and a link to the genome sequence database for S. kunkelii developed at the University of Oklahoma's Advanced Center for Genome Technology in collaboration with the MPPL.
Spiroplasmas were discovered and the term "spiroplasma" was coined by Dr. Robert E. Davis during his research on the corn stunt disease. The helical, motile, wall-less prokaryote was the first known of a large group of unique bacteria. Following Davis's discovery, spiroplasmas were later recognized in at least two other plant diseases, were found in numerous species of insects, and were identified as the causal agents of diseases in fresh water and marine shrimps and in the Chinese mitten crab.
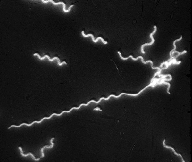
Spiroplasma kunkelii is a helical, cell wall-less prokaryote (bacterium) that causes the plant disease known as corn stunt or achaparramiento. When discovered in the MPPL, it was the first helical, cell wall-less organism known. S. kunkelii is transmitted from corn plant to corn plant by insect vectors, specifically leafhoppers in the genus Dalbulus.
The complete sequence of the genome of S. kunkelii strain CR2-3x is being determined in a collaboration initiated among the USDA-Agricultural Research Service's Molecular Plant Pathology Laboratory (MPPL) and the University of Oklahoma's Advanced Center for Genome Technology (ACGT).
Other participants include the University of Brasilia's Laboratory of Molecular Biology, the Oklahoma State University's Plant Pathology Department, and the Phytovirus Laboratory, Institute of Botany in Vilnius, Lithuania.
