| Enhancement of Crop Plants |

|

Enhancement of Crop Plants for Superior Salinity Tolerance
Global reduction in the availability of clean water in arid and semiarid regions is forcing farmers to irrigate with low-quality degraded/recycled waters, which are generally higher in salts than freshwater. ARS scientists at the Salinity Laboratory are focused on advancing crop plant resilience to salinity by studying the genetic networks that regulate salinity tolerance mechanisms. Their work aims to develop salt-tolerant genotypes that can thrive when irrigated with recycled or highly saline water, enabling crop cultivation on marginal lands. This research is poised to help farmers adapt to water scarcity while maintaining high productivity, ultimately supporting more sustainable agricultural practices.
Research Projects
Developing Salt-Tolerant Alfalfa
Expression Analysis
Alfalfa is a critical forage crop for the dairy industry, valued for its perennial growth, high biomass yield, excellent nutritive quality, and palatability, but with substantial water requirements. Recent studies by Salinity Laboratory scientists on the effects of salinity on growth, biomass yield, photosynthesis, water and ion relationships, nutritive value, antioxidant capacity, and gene expression provided a clear picture of different component traits involved in the salinity response of alfalfa.
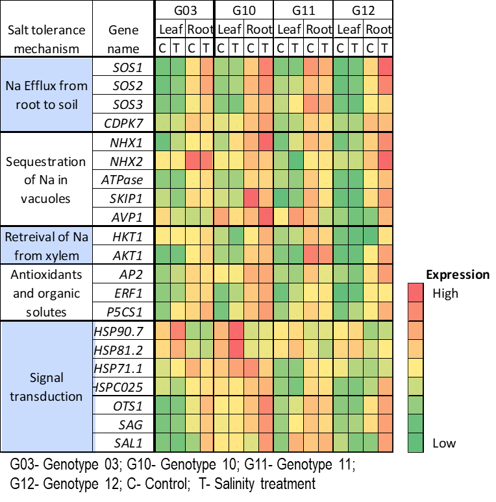
Expression analyses of 21 genes regulating different component traits of salinity tolerance mechanism in four alfalfa genotypes.
Crossing Genotypes Based on Component Traits of Salt Tolerance Mechanism

Dr. Devinder Sandhu is crossing diverse genotypes based on their component traits related to their salt- tolerance mechanism.
Based on gene expression analyses, the genotypes were categorized according to their specific component traits contributing to salinity tolerance mechanisms. A diverse set of genotypes was strategically crossed with the goal of combining multiple salinity tolerance traits into a single, robust genotype. The resulting segregating populations were then evaluated for their salinity tolerance using an outdoor lysimeter system. Individual genotypes were assessed and classified based on total biomass production and ion accumulation under saline conditions, offering a clear distinction of their overall performance and adaptability to salinity stress.
New Alfalfa Genotype That Can Tolerate Seawater Salinity
Selected lines that exhibited high biomass production under saline conditions were further screened for survival at half-seawater salinity. Two lines demonstrated the ability to survive at these elevated salinity levels. The progeny of these lines were then exposed to full seawater salinity, where only a few genotypes survived, demonstrating their exceptional tolerance to extreme saline conditions. ARS scientists at the Salinity Laboratory have a few additional genotypes that are in the pipeline and show extreme tolerance to salinity stress.

A new alfalfa genotype (right) can survive at seawater salinity while the standard cultivars (left) dies even at half seawater salinity.
Characterizing Almond Rootstocks For Salinity Tolerance
Screening Commercial Almond Rootstocks For Salinity Tolerance

Screening of 16 almond rootstocks showed tremendous variation for salinity stress.
Sixteen commercial almond rootstocks were evaluated under five different types of irrigation water to investigate the genetic, physiological, and biochemical mechanisms behind salt tolerance. The treatments included a control group and four saline water treatments, each dominated by different salts: sodium-sulfate, sodium-chloride, sodium-chloride/sulfate, and calcium/magnesium -chloride/sulfate. The water high in sodium-chloride caused the greatest reduction in survival rates and trunk diameter, indicating that sodium (Na) and, to a lesser degree, chloride (Cl) were the most harmful ions to almond rootstocks. Peach hybrids and peach-almond hybrids demonstrated the highest tolerance to salinity.
Leaf Na And Cl Accumulation Negatively Correlates With Salinity Tolerance
The performance of rootstocks under salinity was strongly correlated with their leaf sodium (Na) and chloride (Cl) concentrations, highlighting that the ability to exclude Na+ and Cl− is essential for salinity tolerance in Prunus species.
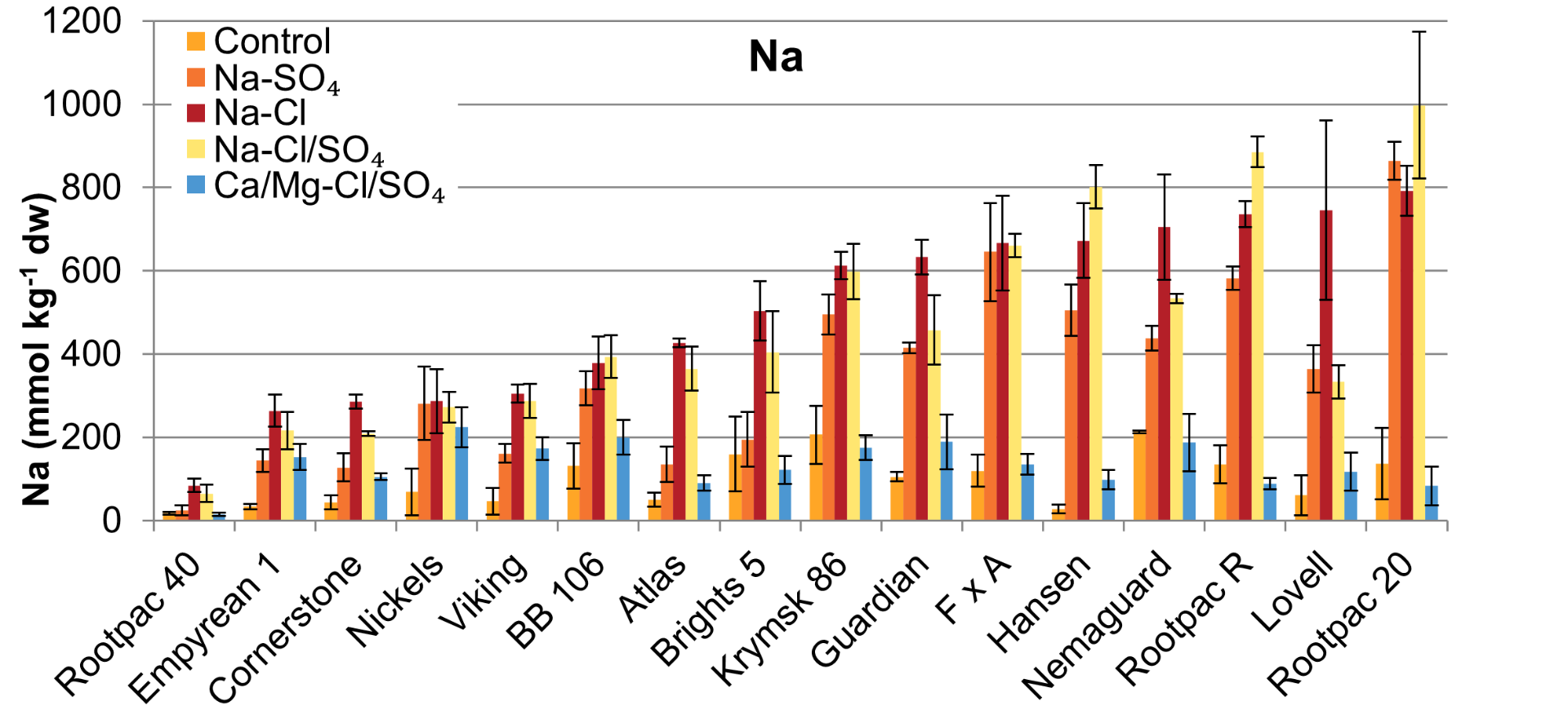
Salt-tolerant genotypes exhibited lower sodium (Na) accumulation in their leaves, while salt-sensitive genotypes accumulated significantly higher amounts of Na
ARS scientists at the Salinity Laboratory are actively collaborating with almond breeders to develop new almond genotypes that exhibit improved tolerance to saline conditions. This collaborative effort, partly funded by the Almond Board of California, integrates cutting-edge genetic, physiological, and biochemical approaches to identify key traits contributing to salt tolerance.
Functional complementation of the Almond HKT1 (PpHKT1) gene in the Arabidopsis hkt1 mutant

Transgenic lines PpHKT1OE2.2 and PpHKT1NP6 expressing PpHKT1 exhibit enhanced salt tolerance compared to the Arabidopsis hkt1 mutant, suggesting that PpHKT1 can successfully restore salt tolerance function in Arabidopsis.
The high-affinity K+ transporter 1 (HKT1) plays a critical role in removing Na+ from the xylem and returning it to the root, thereby mitigating sodium toxicity. ARS Scientists in Riverside complemented the Arabidopsis athkt1 knockout mutant with an HKT1 ortholog (PpHKT1) from the almond rootstock ‘Nemaguard’. Transgenic Arabidopsis lines were developed in the athkt1 background using both a constitutive promoter (PpHKT1OE2.2) and a native promoter (PpHKT1NP6). These lines were exposed to different salt treatments, and both outperformed the athkt1 mutant at 90 mM NaCl, indicating that they successfully restored salt tolerance function in Arabidopsis.
For more information about plant salinity research, contact devinder.sandhu@usda.gov
